Annotations in a model can be stored at multiple levels: the overall model (MIRIAM), model documents (BioModel, MathModel, Geometries), and application, simulations, compartments, species and reactions. Minimal Information Requested In the Annotation of biochemical Models, MIRIAM) functionality in VCell is part of a collaboration in the general modeling community to allow biological models to be "annotated" with a common structure and format describing the components of a mathematical model. A common descriptive format for model elements (molecules, reactions, compartments, etc…) makes understanding models and sharing information easier amongst users. MIRIAM annotation also allows software applications to make more intelligent decisions when importing models created in different environments. File -> Model Annotation allows the user to view/edit MIRIAM annotations for model entities.

Viewing/Editing MIRIAM Annotations
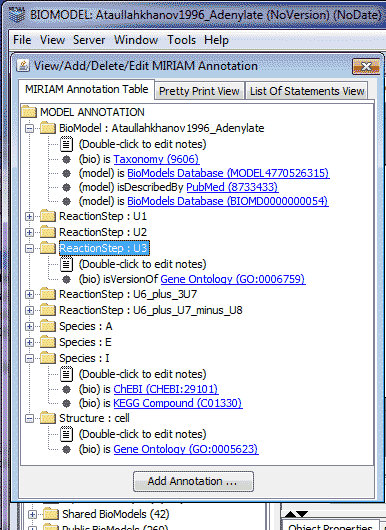
VCell users may view/edit MIRIAM information in BioModels using the MIRIAM editor accessed from File -> Model Annotation menu sequence from any VCell main window. This brings up a window to view/add/delete/edit MIRIAM annotations for a model. Currently, the MIRIAM annotations are viewable for models that are imported from the 'BioModels Database' (http://www.ebi.ac.uk/biomodels-main/) that have MIRIAM annotations. The MIRIAM annotation is now represented in a tree format with a node for each model component (the model itself, reactions, species, and structures). Each node displays the model component and its name. Any other MIRIAM information (its references in common databases) for the model component is displayed as a leaf of the component. Each database reference leaf node is displayed in the following format: Annotation_Scheme, Annotation_Qualifier, Database_name (id_of_component_in_database).
Annotation_Scheme defines the modeling community organization that provides the definition of the terms used to describe a model component. Annotation scheme can be a model qualifier or a biology qualifier.
Annotation_Qualifier lists the predefined (by Annotation Scheme) terms used to describe a model component, for example, ‘is’, isDescribedBy’, isVersionOf, isPartOf, etc.
Database_name is the name of the database from where the reference was found. Some common databases are Gene Ontology, KEGG, ChEBI, UniProt, InterPro, etc. "Id_of_Component_in_database" : represents the id of given component in the database referred to by 'Database_name'. For example, if there is a species in the model for Cyclin-C which is found in the UniProt database, its id in the UniProt database is ‘Q4KLA0’.
Adding Annotations
The user can also choose to add annotation to a BioModel entity (BioModel, reaction, species or compartment/structure) by selecting the entity and clicking on the Add Annotation button in the "View/Add/Delete/Edit MIRIAM Annotation" window. This brings up a Choose Annotation Type dialog. The user has 2 options:
Identifier
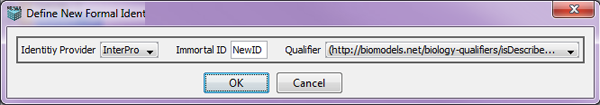
Choosing the "Identifier" option and clicking OK brings up the Define New Formal Identifier dialog. This dialog allows the user to choose an "Identity Provider" from a drop-down list (such as UniProt, Gene Ontology), "Immortal ID" (a new ID) and a "Qualifier" from a set of available qualifiers in a drop-down list.
Date
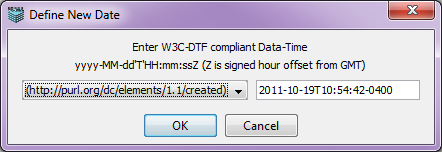
Choosing the "Date" option in the "Choose Annotaion Type" dialog and clicking OK pops up the Define New Date dialog. This dialog allows the user to enter a W3C-DTF compliant Date-Time for the selected entity.
For more information, please refer to MIRIAM.