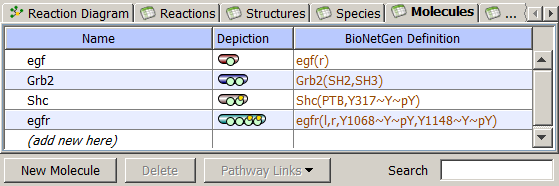
Molecules are the principal component of rule-based model specification. They are used to create Species and to define Reaction Rules and Observables. Molecules are structured objects comprised of sites that can bind other sites, either within a single molecule or between molecules. Sites typically represent physical parts (e.g. domains) of proteins. Sites may also be associated with a list of states, intended to represent states or properties of the site, e.g. phosphorylation status.

New Molecule creates a new molecule with a default name of MT'number', and also creates a new observable with the defaul name of O'number'_MT'number'_tot. The Observable tracks the amounts of all instances of the molecule in all species. (Note that if the Molecule is conserved (i.e. not synthesized or degraded), then the observable Molecule_tot will be conserved, which is one possible verification of model consistency.)
Edit the molecule name in the table; molecular properties cannot be edited in the table, but must be specified in the Molecule Properties pane.
Search the list of molecules by entering a name or an element of molecular structure in the Search box below the table.
Delete molecules by selecting the row and using the Delete button below the table. A molecule will not be deleted if it is used elsewhere in the model.
Add or Edit Annotations using the Annotations tab in the Properties Pane.