Pathway Commons Database
The Pathway Commons panel allows to query and download biological pathways from Pathway Commons in BioPAX Level 2.
Pathway Commons aggregates and integrates the content of nine major pathway databases (BioGRID, MSKCC Cancer Cell Map,
Human Protein Database, HumanCyc, SBCNY IMID, IntAct, MINT, NCI/Nature PID, Reactome).
As of October 2011, Pathway Commons contains 1,668 pathways, 442,182 interactions, 86,282 physical entities and 414 organisms.
Downloaded pathways can be used to build and annotate models based on the pathways and their links to terminologies and databases
about proteins, small molecules, genes, anatomical features, organisms and publications.
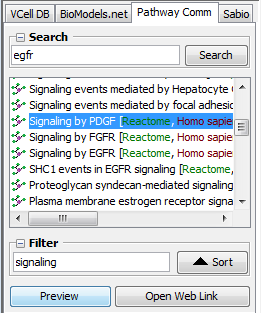
- Search textfield
Enter 1 or more space separated text fragments that will be used to match model names and meta-data information in the Pathway Commons repository.
- Search button Search the Pathway Commons repository using the search text.
- Search list Displays downloadable pathway names (with source and organism name) matching search criteria.
select 1 entry from the list to be downloaded from the remote repository.
- Filter textfield Enter text to match name, database or organism in the current 'Search list' to refine search results.
- Filter Sort button Press to toggle sort of the current 'Search list' by pathway names in ascending or descending order.
- Preview button Press to download the pathway information selected in the 'Search list'.
Once the download is complete a table of pathway subcomponents is displayed under the Pathway Preview tab in the lower right panel of the VCell document window.
See Pathway Preview for information about importing pathway subcomponents into VCell.
- Open Web Link button Press to launch a web browser that displays the Pathway Commons web page of the pathway currently selected in the 'Search list'.

