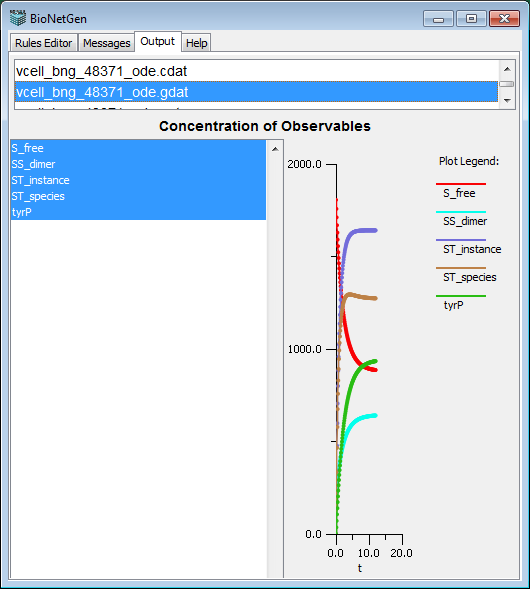
BioNetGen Output tab
The output tab has two fields. In the top text field the user can see the list of all files generated by BioNetGen. When selected, the file is displayed in the bottom text field.
Input BNGL file is included, as well as all BioNetGen outputs. All files have the same prefix vcell_bng_NNNNN, where NNNNN is the random number generated by BioNetGen.
Extension of the file specifies the type of output.
Output types
- .bngl BioNetGen Input file with a set of rules
- .net Biochemical reaction network file generated by BioNetGen generate_network{} command, in a short BNG format
- _end.net Biochemical reaction network at the end time point of network simulation
- .cdat Output file with a tab-delimited set of concentrations for all species. It is
a result of simulation using simulate_ode{} and simulate_ssa{} commands
- .gdat Output file with a tab-delimited set of concentrations for all observables (user-specified functions). It is
a result of simulation using simulate_ode{} and simulate_ssa{} commands
- .xml SBML output file with the biochemical reaction network. Depending on the position of
writeSBML() command it can be either an initial network or a network at the end time point of simulation
- .m MatLab output file with the biochemical reaction network. Depending on the position of
writeMfile() command it can be either an initial network or a network at the end time point of simulation
- .tex LaTeX output file with the biochemical reaction network - differential equations only
On the right of the tab there are two buttons:
- Save output as a text file - Click to save the selected file to your computer.
- Create a BioModel - This button is enabled when .xml file is generated. Clicking on this button launches a
VCell window with the BioModel corresponding to this .xml file.

