VCML is the Virtual Cell Markup Language and the native Virtual Cell format. The Virtual Cell supports interoperability with other standard representations for biological systems modeling. Non-spatial BioModels can be easily exported as XML and Matlab files when users want to use their models outside of the VCell environment.
Note: If you plan on importing the model back into the Virtual Cell, make sure to export a copy as VCML. SBML is an XML format that should only be used if you need to import the models into another program that requires this format.
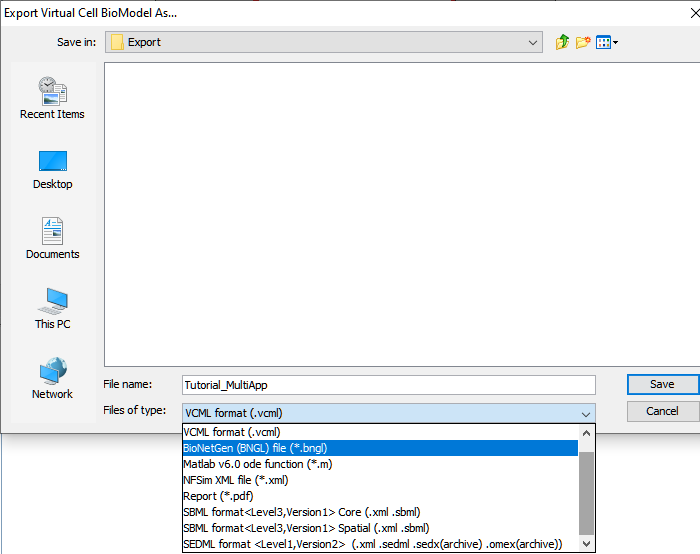
Export Formats Available

To export a model, open the BioModel or MathModel document to be export and choose the File -> Export menu option. This opens a "Choose File" dialog. Navigate to the folder where the model has to be saved on your local drive, select the desired export format from the "Files of Type" combobox and press Save.
Exporting to SBML
SBML export is available only for BioModels. Exporting MathModels to SBML will be provided in future versions of VCell. SBML only supports non-spatial, deterministic modeling. Also, since there is no formal representation for electrophysiological models in SBML, if the BioModel to be exported is electrophysiology based, it is simply not exported and an error message is displayed.
Multiple non-spatial applications
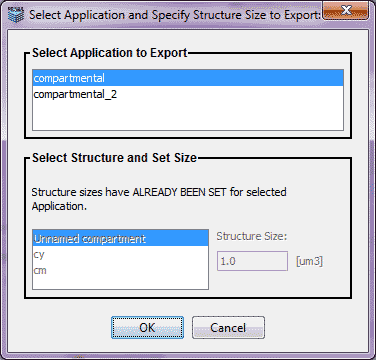
When a VCell BioModel has multiple applications, a list of the non-spatial, deterministic applications is presented. Select the application to export to SBML. If the compartment sizes for the selected application are not already set, the user is asked to provide the size for any one compartment in the BioModel.
Mapping Structures
SBML requires absolute sizes for compartments. If in the selected application, compartment sizes have not been set (i.e. in models generated prior to VCell Version 4.4), the user is prompted to provide the size of any one compartment. Compartment size units are um3 for feature and um2 for membrane. All compartment sizes will be automatically computed using a constraint solver, the user input and relative sizes of the compartments (volume fractions and surface to volume ratios).
The panel to set compartment size is disabled if the sizes of all the compartments have been previously set. If a model was generated with VCell 4.4 or later, all sizes for non-spatial applications are already in the required absolute terms.
Unit Conversions
While exporting to SBML, Virtual Cell converts species concentrations to 'molecules per um3' or 'molecules per um2', volume to 'um3', area to 'um2' and kinetic reaction rates to substance/time units (molecules/sec). Due to this conversion process the user may see additional parameters associated with each reaction or species. One such factor is 'KMOLE' or sometimes simplified to '0.00166112956810631', or '602.0' (1/KMOLE) in the expressions (KMOLE is a dimensionless flux conversion factor used internally within the Virtual Cell, with a value of 1/602 uM.um3/molecules).
Parameter overrides
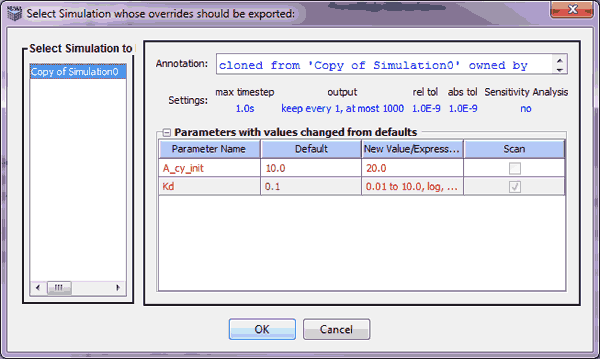
If simulations do not have parameter overrides in the selected application, the selected application is exported to SBML with its default parameter settings. If a selected application has simulations with overridden parameters (parameters changed through the simulation task panel), the user is provided a list of these simulation names and the corresponding "Simulation Summary" is displayed. The user can select one of these simulations and press OK. The selected simulation (with parameter overrides) is exported to SBML. If the overridden parameter is a parameter scan, each scan value is exported into a different SBML file. The names of the exported parameter scans will be of the form: exportFileName_Indx.xml. The 'exportFileName' is the user-provided name for the exported file. 'Indx' is the number of the parameter scan.
For example, if the simulation has a parameter 'P' with 5 scan values, 5 different SBML files are generated. each file corresponds to each scan value of 'P' and 'Indx' takes on the values 0, 1 ...4;. Pressing Cancel will ignore the parameter override simulations and export the default parameter values of the chosen application to SBML.
Expressions in Initial Conditions
If the BioModel to be exported contains species that have initial conditions specified as an expression, such species initial conditions are exported to SBML as an assignment rule in SBML Level2 and Level 3 ONLY IF the species is marked as "Clamped".