NFSim Simulations Editor
Integrator is NFsim biochemical reaction simulator
designed to stochastically simulate systems that have a large or even infinite number of
possible molecular interactions or states. A publication describing NFsim can be
found at https://www.nature.com/nmeth/journal/v8/n2/full/nmeth.1546.html.
A user can set end simulation time, output interval N (simulations results are outputted every N seconds)
and Advanced Solver Options.
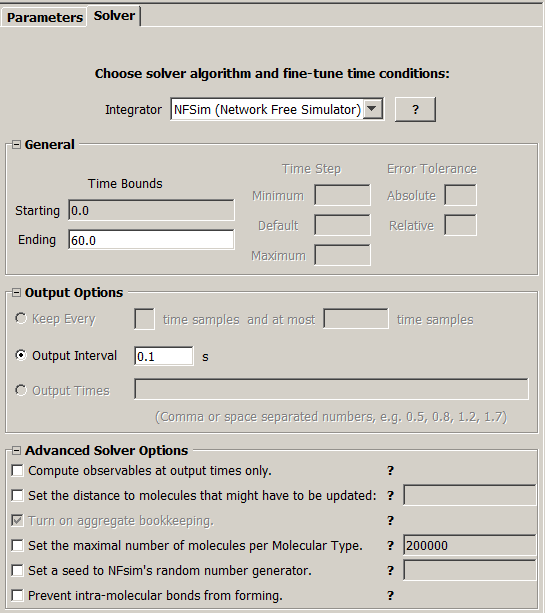
Advanced Solver Options
- Compute observables at output times only (NFSim option -notf).
By default, observables are calculated on-the-fly, updating all observables at each simulation step.
This is necessary when rates of reactions depend on Observables. By checking this box,
the values of Observables will be computed only at output steps. It can not be set if
functions are used, as functions rely on having updated Observables at any point in a
simulation.This will allow simulations to run faster if the number of simulation steps
between each output step is greater than the number of molecules in the system. This
may or may not be true for your simulation, so you should try turning on or off this
option to see which is more efficient.
- Set the distance to molecules that might have to be updated (NFSim option -utl [integer]).
The universal traversal limit (UTL) sets the distance neighboring molecules have to be to the
site of the reaction to be updated. The default UTL is set to the size of the largest reactant pattern,
which is guaranteed to produce correct results because NFsim will always find the changes that apply
to every reactant pattern in the system. Sometimes however, based on the structure of the reactant
patterns, the UTL may be set lower. The lower is the UTL, the less molecules will be checked and
the faster simulation will go. If the limit is too low, not all molecules are correctly being updated,
then results will be incorrect. In many cases, however you may have a very large pattern, but the
maximal number of bonds you need to traverse to make sure that pattern can always be matched is low.
This will happen, for instance, when many molecules are connected to a single hub molecule.
Default: the size of the largest reactant pattern in the rule-set.
- Turn on aggregate bookkeeping - always ON. (NFSim option -cb)
NFsim by default tracks individual molecule agents, not complete molecular complexes.
This is useful and makes simulations very fast, but is not always appropriate.
For example, in some systems it is necessary to block intra-molecular bonds
from occurring to prevent unwanted ring formation. However, to check for
intra-molecular bonding events, complete molecular complexes must be traversed.
NFsim, provides an aggregate bookkeeping system for molecular complexes that form
by assigning each connected aggregate a unique id. Then, it becomes easy to check
if any two molecules are connected. The trade-off is that there is an overhead
involved with maintaining the bookkeeping system with a cost that depends on
the size of the molecular complexes that can form. Default: ON, because VCell
generates observable for every species.
- Set the maximal number of molecules per Molecular Type. (NFSim option -gml [integer])
To prevent a computer from running out of memory in case too many molecules are created,
NFsim sets a default limit of 200,000 molecules of any particular Molecule Type from being created.
If the limit is exceeded, NFsim just stops running gracefully, thereby potentially saving your
computer. NDefault: 200,000
- Set a seed for NFSim's random generator. (NFSim option -seed [integer]) Provide a seed to NFsim's random number
generator so exact trajectories can be reproduced. If this line is not entered,
the current time is used as a seed, producing different sequences for each run.
- Prevent intramolecular bonds from forming. (NFSim option -bscb)
Block same complex binding throughout the entire system. This prevents intra-molecular bonds
from forming, but requires complex bookkeeping to be turned on.

