A Network-Free simulation (using the NFSim solver) can be run for BioModels that include only a single compartment and where all reactions in the model have mass action kinetics. For Biomodels without rules or molecular definitions, the simulations results will generate equivalent results as when using a non-spatial stochastic solver. The Network-free Specifications tab is visible only in Network-Free Applications. When a NetworkFree simulation is run for a non-ruled based BioModel, the Physiology is first coverted into a rule-based model and the simulation is run using the NFSim solver. Within this Specifications window, it is possible to save the rule-based model that will be created as a new VCell Biomodel.
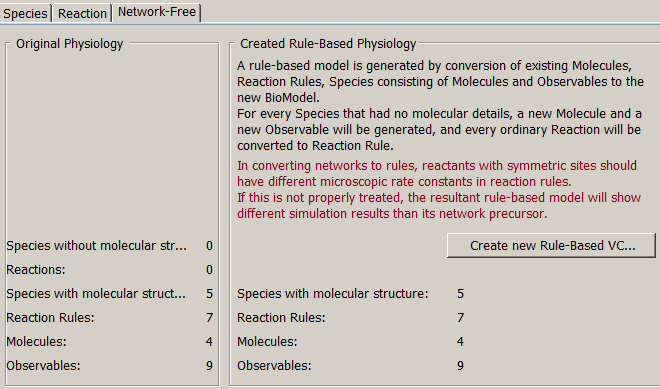
Statistics of the species, molecules, reactions and observables are provided for both the original physiology and the created rule based physiology. If the original model is rule-based, these numbers will be identical.

Create New Rule-Based VCell BioModel from the original physiology. A new BioModel is created and opened in a new window. A rule based model can be created only for models with a single compartment and if all reactions have mass action kinetics. Molecules, observables, species with molecular details, and reaction rules are transferred to a new BioModel without changes. Species without molecular details and reactions are converted into rule-based physiology by generating a molecule for each species, and converting reactions to fully-defined reaction rules.
Note that the same reaction expression treated as a reaction and as a reaction rule will have different rates: