HDF5 files exported from spatial simulation results, can be viewed using either a dedicated HDF5 viewer (see https://www.hdfgroup.org/downloads/hdfview/)or FIJI/ImageJ (see https://imagej.net/software/fiji/). Instructions for importing HDF5 files into ImageJ is given here.

HDF5 files (*.hdf5) exported from spatial simulations
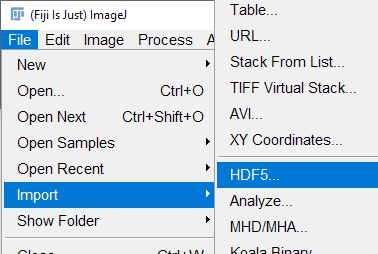
Data are saved as multi-dimensional arrays in a heirarchical xyzt format defined by the HDF5 standard. To open the files in ImageJ, from the File menu select "Import" then "HDF5"
Select all of the datasets ending in ".../DataValues (XYZT)" to be imported. If you exported data for multiple variables in a single HDF5 file, the data for each variable appears as a separate data set path in the table.
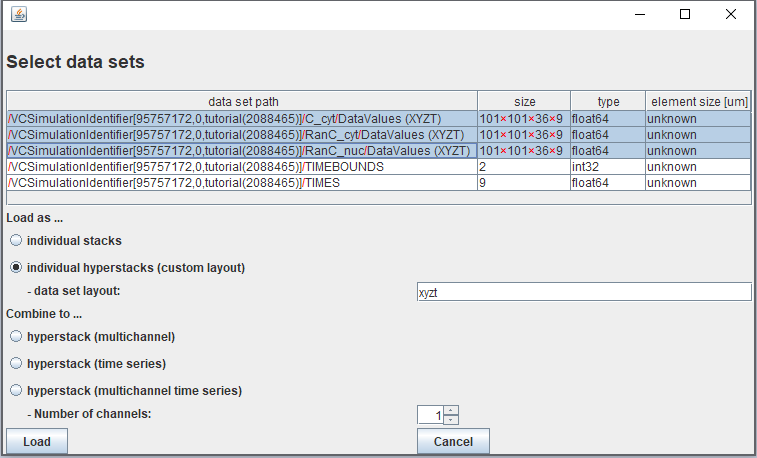
Click "Individual Hyperstacks (custom layout)" and enter "xyzt" in the text box. (Note that it is not yet possible to use the "Combine to..." functions with the exported VCell HDF5 files.) Then select "Load"
The hyperstack viewer with the image will appear. If multiple files were selected, each will appear in a separate image window; the variable represented by each image will be at the end of the rather long filename for the hyperstack datset. Note that the distance scales will not be imported and should be manually changed ("Image"->"Properties") to accurately reflect the mesh size (x,y, and z) in the simulation.
Pixel values in the image are the actual concentration values from the simulation. You will need to adjust the image brightness to correctly display the image (Image->Adjust->Brightness/Contrast).
The images will be rotated and flipped compared to the image view in the VCell databrowser. To align with the VCell viewer, use "Image->Transform->Flip Vertically" and "Image->Transform->Rotate 90 degrees right" functions in ImageJ.