Observables Table
The observables are used to specify model outputs, which are functions of the
population levels of multiple chemical species that share a set of properties. For example, to
simulate the tyrosine phosphorylation level of a particular protein, one need to specify
Observable that is a function for the total amount of all chemical species containing
the phosphorylated form of this protein.
- Each Observable consists of one or more Species Patters. Observable selects species that match at least one
of the species patterns (not both).
- Each species pattern defines common features of species. It consists of one
or more of molecules that compose
species. Molecules may or may not be explicitely connected, but must be present
in every species selected by this species pattern. Sites that have possible multiple states have yellow sign above site shape.
- Each molecule within species pattern may have some sites being undefined. If site
does not have defined states (if states are possible) and its binding is not important, it is shown in white.
- Sites may have defined binding - they can be declared to be explicitly bound to another site,
declared unbound or declared bound to "some" (unspecified) site - then the site is shown in green with bond (if exists)
shown in solid color.
- Site than has defined state is always shown in green.
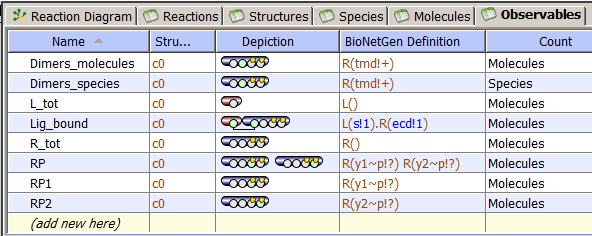
The Observables Tab displays
- Name of Observable (editable).
- Structure where species selected by Observable are located
(currently, a single compartment only).
- Depiction of each Observable. Green circle within each molecular shape denote
molecular sites, small yellow circles denote that this site has possible states.
-
BioNetGen definition that is unediatable but searchable.
- Count either Molecules or Species. In the first case, the output is
a weighted sum of the population levels of the chemical
species matching the pattern(s) in the observable. Each population level is multiplied by the number of times that the species is
matched by the pattern(s). For example, a species that has two instances of a receptor is counted twice.
In the second case, the output is an unweighted sum of
the population levels of the matching chemical species.
For example, a species that has two instances of a receptor is counted once.
Edit An Observable name can be specified and edited in a table, but definition and properties
must be specified in
the Observables Properties bottom pane.
Filter the list of observables using the Search Box to enter a name or any part of observable composition.
Delete observable by highlighting and using the "Delete" button below the table.
Add annotation about the observable within the
Observable Properties pane in the bottom window.
