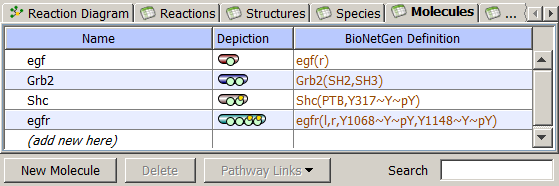
Molecules are the principal component of rule-based model specification. They are used to create Species, define Reaction Rules and Observables. Molecules are structured objects comprised of components (called sites) that can bind to each other, both within a molecule and between molecules. Sites typically represent physical parts of proteins, such as the SH2 and SH3 domains of the adapter protein Grb2. Sites may also be associated with a list of states, intended to represent states or properties of the site, e.g. phosphorylation status
The Molecules Tab displays a table of the Molecule name, Depiction of each molecule and the
BioNetGen definition that is unediatble but searchable.
In the Cartoon, green circle denotes
molecular site, while small yellow circle denotes that this Site has possible states.
Selecting molecule brings the Molecule Properties
in the bottom right pane, where a user can specify and edit a selected Molecule.

New Molecule Once a molecule is entered here, a new observable with a name Molecule_tot is defined. It counts amounts of all instances of this molecule in all species. If the Molecule is conserved (not synthesized or degraded), then the observable Molecule_tot will be conserved - it is one possible verification of model consistency.
Edit A molecule name can be specified and edited in a table, but molecular properties must be specified in the Molecule Properties bottom pane. Molecule edited here is updated everywhere in the model - in Species, Reactions and Reaction Rules and Observables.
Search the list of molecules by entering a name or an element of molecular structure below the table.
Delete molecules by highlighting and using the "Delete" button below the table. Molecule will not be deleted if it is used elsewhere in the model.
Add annotation about the molecules within the Molecule Properties pane in the bottom window.