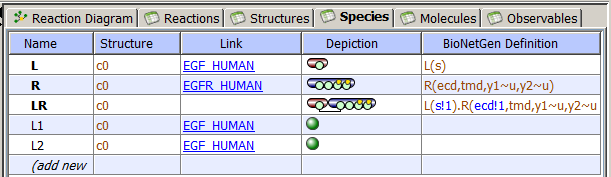
Species are used to create variables (molecular species) in reactions and fluxes. Species can exist in both membranes and compartments (volumes) of the cellular structures. Species are unique pools of molecules within a single compartment. If the same species is copied to different compartment, the compartment name is appended to the species name to retain a unique identifier, e.g. species_compartment. Species is shown in Reaction Diagram as a green shape.
For use in rule-based modeling species must have molecular details specified: they are composed of molecules with bonds connecting binding sites. If Molecule has multiple states, each state must be specified - otherwise the species will present a pool of different chemical entities and will not be valid. Species with molecular details can be used as regular species in any reaction network, but they also can serve as seed species in rule-based models: they are either used as starting set of species to generate a network (using BioNetGen software), or used as an initial set of species for stochastic network-free simulations performed by (using NFSim software).

The Species Tab has several columns:
Edit molecular details of species in the Species Properties pane in the lower window.
Filter the list of species using the Search Box to enter a species name or a structure name.
Delete species by highlighting and using the "Delete" button below the table.
If you wish to use Pathways Commons data, select the species and use the Pathway Links button.
Add annotation about the species within the Species Properties pane in the bottom window.