Network Specifications
For
deterministic and
stoichastic applications,
Network Tab describes constraints for network-generation performed by
BioNetGen engine, and tests
the size of the reaction network that is obtained using these constraints.
Only after the user applies changes, the simulation
is performed on the generated reaction network.
This tab is active only if at least one Molecule is specified.
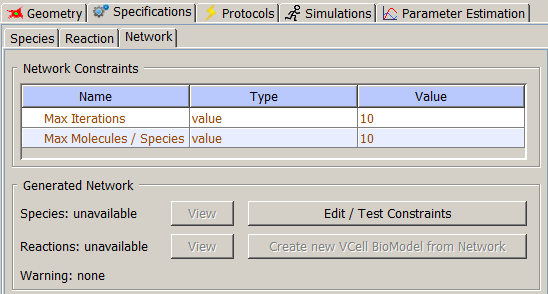
- Max Iterations - how many times the rules are applied to the set of
seed species to generate new species and reactions.
- Max Molecules/Species - the maximal number of molecules of any type that can be in a single species.
BioNetGen will not apply any rules to species reaching this limit.
- Species - the total number of species in the reaction network generated under
specified constraints, including seed species.
- Reactions - the total number of reactions in the reaction network generated under
specified constraints.
After clicking on "Edit/Test Constraints" button, a user is presented with a window to specify new constraints.
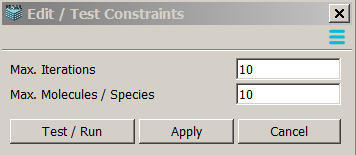
- Test/Run will attempt to generate a reaction network using these constraints.
- Apply will apply these constraints without network generation,
so the simulation will be performed sequentially after network generation.
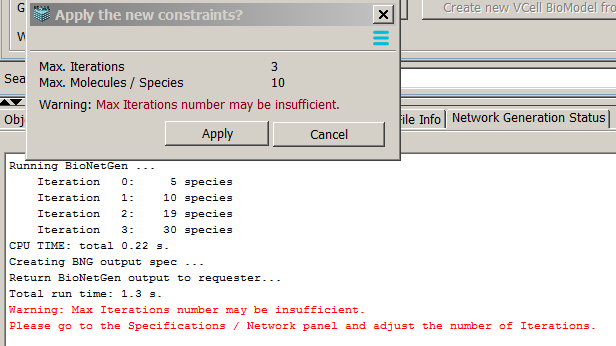
Running network may generate some warnings:
- Max iterations number may be insufficient means that the network may be not exaustively generated. In the
Network Generation Status pane below one can see that the number of species during the last two iterations is
different. Normally, the network generation stops when the last iteration does not produce new species, which means that
the number of species at the last two iterations are the same.
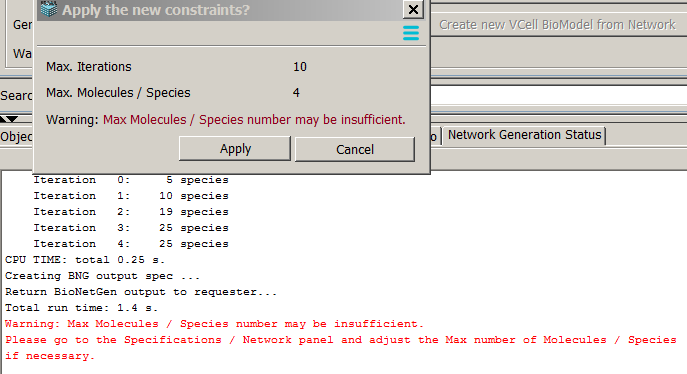
- Max Molecules/Species number may be not sufficient means that the species with the set number of molecules are still
subject to rules application, so the network is truncated by this limitation.
 Note: When the warning is displayed, the network is trunkated, which means that NFSim simulation will
likely produce different results than deterministic or stochastic simulations that use this network.
Note: When the warning is displayed, the network is trunkated, which means that NFSim simulation will
likely produce different results than deterministic or stochastic simulations that use this network.
When Apply button is pressed, the constrains are applied to the application, and the user can
View all generated species and reactions, as well as Create a new VCell BioModel that consist
of all generated species and reactions. This new model is not rule-based, but all species have molecular detail spe specified.
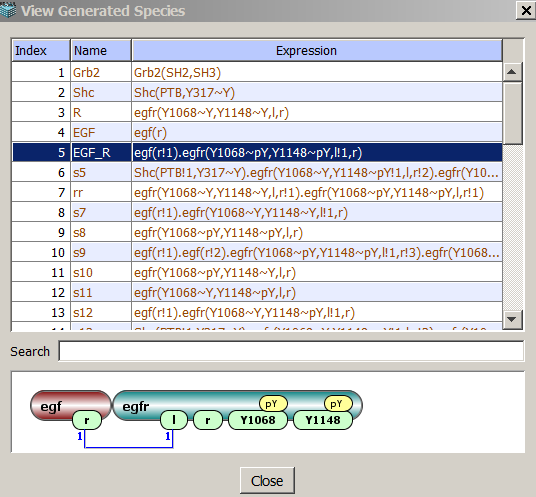
- View Generated Species/View Generated Reactions After constraints are approved by the user,
species and reactions are generated and can be viewed by pressing View button.
The pop-up window shows species name, depiction and searchable BNGL code. Species names are the same as
seen when checked "Generated Species" in "Simulation Results" window.
- Create New VCell BioModel from Network launches a new BioModel in a new window.
This BioModel consists of all generated species and reactions, species have molecular details,
while reactions carry rule name under reaction name.
Note: reaction rates in reaction network are adjusted for
symmetry factors and
statistical factors.




 Note: When the warning is displayed, the network is trunkated, which means that NFSim simulation will
likely produce different results than deterministic or stochastic simulations that use this network.
Note: When the warning is displayed, the network is trunkated, which means that NFSim simulation will
likely produce different results than deterministic or stochastic simulations that use this network.
