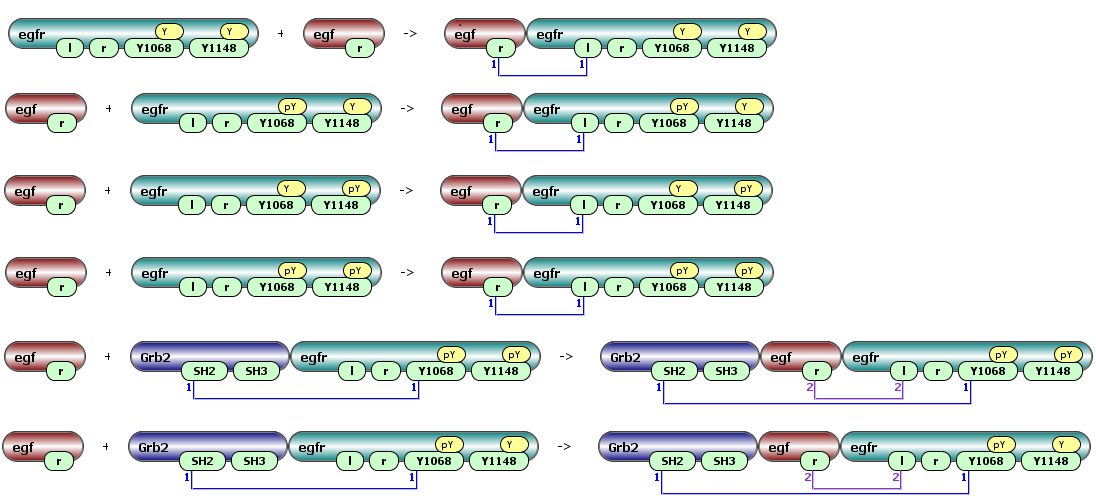
A rule-based model is specified as a set of reaction rules, which are associated with specific rate laws. Given a set of seed species, a reaction rules identifies those species that have the features required to undergo the transformation from reactants to products specified in the reaction rule. Interactions represented in a reaction rule do not depend on features not explicitly indicated. Thus, multiple species may qualify as reactants in a type of reaction defined by a reaction rule.
The modeler can define which components and modifications of a molecule or molecular assembly affect a particular chemical transformation, and which do not. Furthermore, the modeler has the ability to account for steric clashes, cooperativity, and any other factors that might influence the rate of a reaction. A reaction rule can state, for example, that “any cell-surface monomeric receptor having an available extracellular binding site and any free extracellular ligand can interact and form a ligand–receptor complex; the probability of this interaction depends only on the total numbers of cell-surface monomeric receptors and extracellular ligands and does not depend on the specific state of the receptor cytosolic portion.” In this example, we assume that the cytoplasmic state of a receptor does not affect ligand–receptor binding, which implies that to parameterize all reactions specified by the ligand–receptor interaction rule, we need just two rate constants: on and off rates.
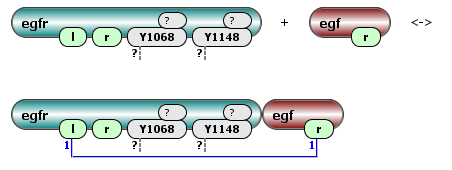
Some of possible reactions generated by this rule: