2018-3-15 Announcing the release of VCell 7.0. VCell 7 includes new 2D kinematics functionality for solving reaction diffusion equations within moving boundaries. To support models of cell motility and morphogenesis, this allows users to specify velocities of points, surfaces and volumes within the geometry. Other features in VCell 7.0 are methods for creating simulated fluorescence for direct comparison to microscopy data, the ability to use COMSOL Multiphysics solver (requires a local COMSOL license) and adaptive meshes for simulating with different spatial scales using EBChombo. VCell 7.0 has also been redesigned with new reusable modules and a cloud-hosted software development processes supporting external collaboration and extensibility (GitHub, Travis-Cl, DockerHub). VCell 7.0 also includes reproducible and portable server and solver deployments using container technologies.
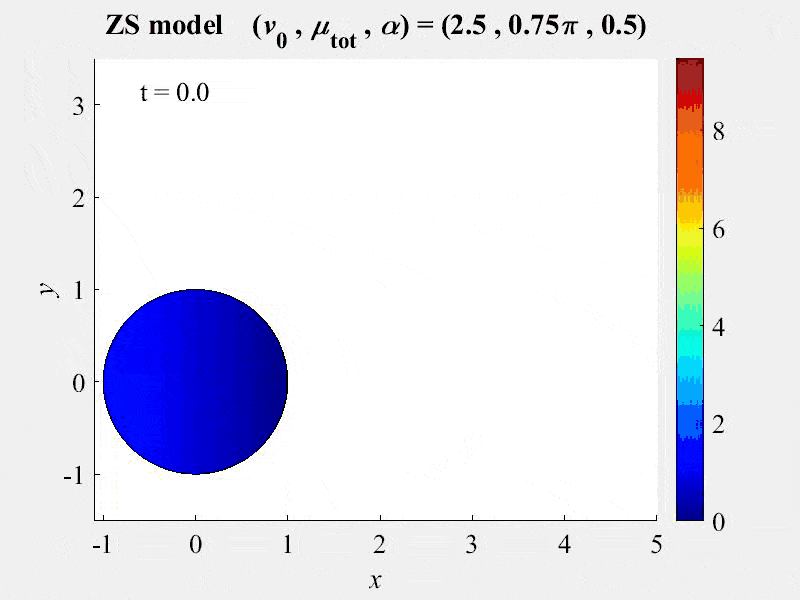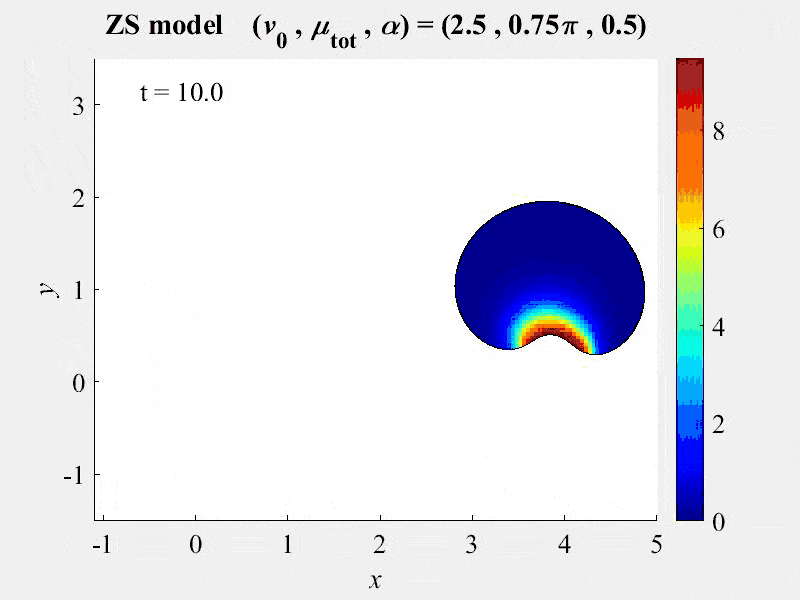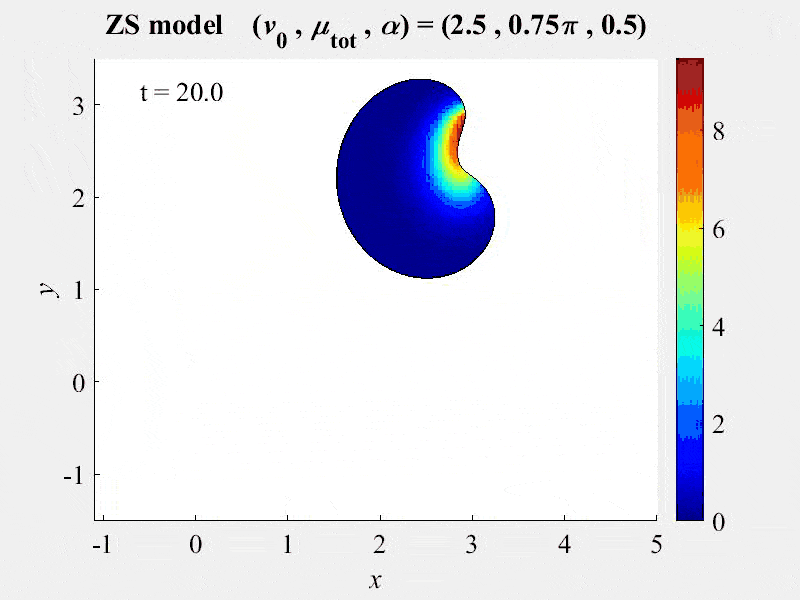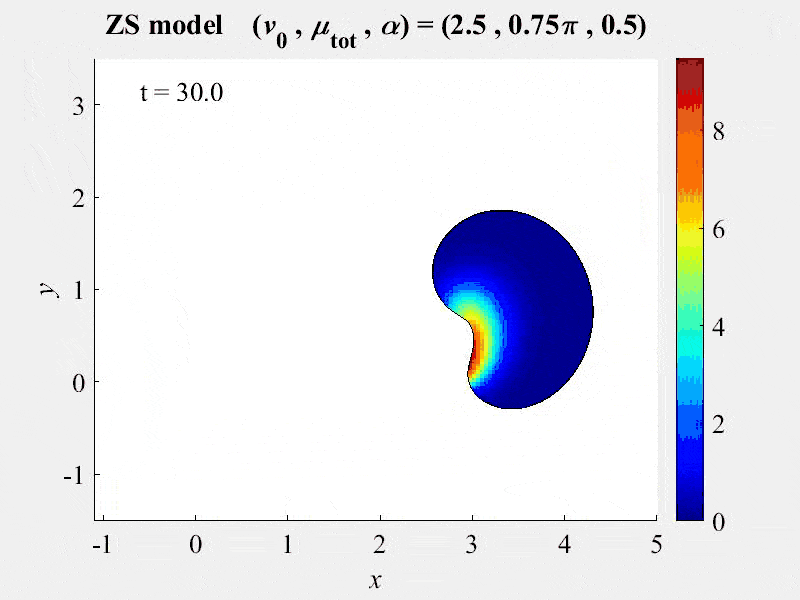
The movie shows cell rotations in a model of actomyosin motility using the kinematics algorithm deployed in VCell 7.0; pseudocolors are myosin concentration. This model also includes cellular mechanics using an algorithm under development for future deployment in VCell. This work, and details of the model, are described in:
M. Nickaeen et al. (2017) A free-boundary model of a motile cell explains turning behavior. PLOS Computational Biology 13(11): e1005862. https://doi.org/10.1371/journal.pcbi.1005862