Events
News
A rule-based VCell model used in J Immunol publication
2025-03-09. Alberto Millan from Kite Corp and Lewis Lanier from UCSF published a study on ITAM-mediated human Natural killer cells signaling. VCell was used to create a rule-based model in .bngl (BioNetGen) format.
Millan, A. J., Allain, V., Nayak, I., Libang, J. B., Quijada-Madrid, L. M., Arakawa-Hoyt, J. S., … & Lanier, L. L. (2025). SYK negatively regulates ITAM-mediated human NK cell signaling and CD19-CAR NK cell efficacy. The Journal of Immunology, vkaf012
https://pubmed.ncbi.nlm.nih.gov/40073103/
The 26th Annual CompCellBio workshop
2025-02-28. The 26th Computational Cell Biology workshop took place ONLINE for the 5 days, February 24-28th. Cell biologists and biophysicists leaned how to use VCell (https://vcell.org/) and SpringSalad (https://vcell.org/ssalad) software to develop spatial and non-spatial models using deterministic, stochastic, agent- and rule-based approaches.
VCell at CellBio 2024
2024-12-18. VCell was represented at the American Society of Cell Biology Annual Meeting 2024 in San Diego. Michael Blinov and Margaret Johnson (John Hopkins University) led the session on Biophysical Modeling of the Cell on December 14th. On December 15th, Leslie Loew, Ann Cowan, Stephan Hoops (U Virginia) taught COPASI and VCell tutorials during the workshop on
Mathematical Modeling for Cell Biology.
A VCell spatial model of Myosin II and cortexillin I assembly was published by Iglesias team
2024-11-08. The team of researchers from John Hopkins University used spatial modeling with VCell to analyze the role of Myosin II and cortexillin I assembly within the mechanoresponsive mechanism grounded in experimentally measured parameters.
Dolgitzer, D., Plaza-Rodríguez, A. I., Iglesias, M. A., Jacob, M. A. C., Todd, B., Robinson, D. N., & Iglesias, P. A. (2024). A continuum model of mechanosensation based on contractility kit assembly. Biophysical Journal. Nov 8:S0006-3495(24)00708-2.
https://pubmed.ncbi.nlm.nih.gov/39521955/
Computational Cell Biology workshop
2024-07-22-24. VCell and COPASI teams run 25th Annual Workshop on Computational Cell Biology. Nine students (one could not attend due to flights being cancelled) from Arkansas, Connecticut, New York, Ohio, Texas and Viginia worked with instructors for 3 days, developing their computational models. Thanks to NIH for the funding!
.
Reaction-diffusion modeling with VCell used to study biophysical carbon concentrating mechanisms in land plants
2024-06-10. The team of researchers from Michigan State University used reaction-diffusion modeling with VCell to provide insights into biophysical carbon concentrating mechanisms in land plants.
Kaste, J. A., Walker, B. J., & Shachar-Hill, Y. (2024). Reaction-diffusion modeling provides insights into biophysical carbon concentrating mechanisms in land plants. Plant Physiology, kiae324.
https://pubmed.ncbi.nlm.nih.gov/38857179/
A VCell model of endocytic nanoclusters is published in Nature Comm
2024-05-14. The team of researchers from Australia and Johns Hopkins University used VCell to design and simulate a model explaining the formation of endocytic nanoclusters driven by distinct recruitment of Dynamin1 long- and short-tail isoforms.
Jiang, A., Kudo, K., Gormal, R.S., Ellis, S., Guo, S., Wallis, T.P., Longfield, S.F., Robinson, P.J., Johnson, M.E., Joensuu, M. and Meunier, F.A., 2024. Dynamin1 long-and short-tail isoforms exploit distinct recruitment and spatial patterns to form endocytic nanoclusters. Nature Communications, 15(1), p.4060.
https://pubmed.ncbi.nlm.nih.gov/38017688/
VCell at HARMONY 2024
2024-04-10. Michael Blinov and Ion Moraru presented VCell tutorial at HARMONY 2024 at University College London. Other topics discussed include interoperability of multiple standards like SBML, SED-ML and COMBINE archive supported by VCell.
25th Annual Comp Cell Bio workshop
VCell, COPASI and SpringSalad were taught and used for projects at the 25th Annual Computational Cell Biology (CCB) workshop that took place online February 26-28th. The invited talks were given by Yulia Timofeeva (University of Warwick), James P Sluka (Indiana University) and Kevin Janes (University of Virginia). Sixteen participants were selected to work with VCell and COPASI instructors on their projects, and eight gave presentations on their projects at the Modeling Symposium.
A model of K+ and pH homeostasis in plant cells.
2024-02-12. Ingo Dreyer (U Talca, Chile) and Kai Konrad (U Wuerzburg, Germany) used VCell to design and simulate a model explaining the properties of transporter networks and the coupling of transport across the PM and VM. They demonstrated fundamental principles of coupled ion transport at membrane sandwiches to control H+ /K+ homeostasis and points to transceptor-like Ca2+ /H+ -based ion signaling in plant cells.
Kunkun Li, Christina Grauschopf, Rainer Hedrich, Ingo Dreyer, Kai R Konrad (2024). K+ and pH homeostasis in plant cells is controlled by a synchronized K+ /H+ antiport at the plasma and vacuolar membrane. New Phytol. 241(4):1525-1542.
https://pubmed.ncbi.nlm.nih.gov/38017688/
VCell biomodels is available at https://vcell.org/biomodel-270051643
Special Interest Group on Biophysical Modeling of the Cell
Michael Blinov and Margaret Johnson from John Hopkins University were hosting the Special Interest Subgroup session on Biophysical Modeling of the Cell at Cell Bio 2023 – an ASCB/EMBO (https://www.ascb.org/cellbio2023/) meeting in Boston, MA, December 2-6. The speakers include Elizabeth Read (UCI), Margaret Johnson (JHU), Jason Haugh (NCSU), Graham Johnson (Allen Institute), and Ben O’Shaughnessy (Columbia U).
VCell at ICSB 2023
VCell was highlighted at the 22nd International Conference on Systems Biology (ICSB) that took place in Hartford October 8-12, 2023. The meeting was organized by CCAM faculty and co-chaired by Pedro Mendes and Ion Moraru. VCell team presented two tutorials (basic modeling by Ann Cowan and advanced features by Michael Blinov) and a poster.
A model of the cadherin-11 and PDGFR pathways crosstalk
2023-09-22. VCell collaborator Aurélie Carlier from Maastricht University published a manuscript where a computational model in VCell was used to represent the experimentally proven interactions between cadherin-11 and the two PDGFRs. The authors demonstrated the existence of a crosstalk between β-catenin (downstream to cadherin-11) and an ERK inhibitor protein (e.g. DUSP1).
Karagöz, Z., Passanha, F. R., Robeerst, L., van Griensven, M., LaPointe, V. L., & Carlier, A. (2023). Computational evidence for multi-layer crosstalk between the cadherin-11 and PDGFR pathways. Scientific Reports, 13(1), 15804. https://pubmed.ncbi.nlm.nih.gov/37737289/
VCell biomodels is available at https://vcell.org/karagoz-2023
19 new VCell models on G-actin diffusion are published by Haugh group
2023-08-28. VCell collaborator Jason Haugh from North Carolina State University published a manuscript where experiments and VCell modeling demonstrated that G-actin diffusion is insufficient to achieve F-actin assembly in fast-treadmilling protrusions.
Appalabhotla, R., Butler, M.T., Bear, J.E. and Haugh, J.M. G-actin diffusion is insufficient to achieve F-actin assembly in fast-treadmilling protrusions. Biophys J. 2023 Aug 28;S0006-3495(23)00553-2. doi: 10.1016/j.bpj.2023.08.022. https://pubmed.ncbi.nlm.nih.gov/37644720/
See 5 biomodels and 14 mathmodels at https://vcell.org/appalabhotla-2023
Biophysical Modeling of the Cell special interests group at ASCB
Prof Margaret Johnson from John Hopkins University and Michael Blinov are hosting the Special Interest Subgroup session on Biophysical Modeling of the Cell at Cell Bio 2023 – an ASCB/EMBO (https://www.ascb.org/cellbio2023/) meeting in Boston, MA, December 2-6. The session will take place on Wednesday, December 6, 8:30 AM – 11:00 AM (https://www.ascb.org/cellbio2023/program/subgroups/).
24th Annual Computational Cell Biology (CCB) workshop
2023-06-28. The 24th Annual Computational Cell Biology (CCB) workshop took place at the Center for Cell Analysis and Modeling (CCAM) on June 26-28th. 11 participants from all over the US (from Maryland to Illinois) came to Farmington to work on developing their modeling projects using VCell and COPASI software tools, designed and maintained at CCAM. The students were helped by a CCAM team consisting of Michael Blinov, Ann Cowan, Leslie Loew, Pedro Mendes, Ion Moraru, Kelvin Peterson, Jim Schaff, Nathan Schaumburger, and Boris Slepchenko.
VCell is used in a research on aging
2023-05-04. The interdisciplinary team consisting of CCAM modelers Drs. Blinov and Moraru and researchers from the Center on Aging Drs. Kuchel and Kositsawat, together with former undergrad students Schaumburger and Pally published a manuscript theoretically explaining the bistability of clinical outcomes: the likelihood of an individual remaining mobile over time either increases to almost 100% or decreases to almost zero. The VCell software was extensively used for parameter scans and fitting.
Schaumburger, N., Pally, J., Moraru, I. I., Kositsawat, J., Kuchel, G. A., & Blinov, M. L. (2023). Dynamic model assuming mutually inhibitory biomarkers of frailty suggests bistability with contrasting mobility phenotypes. Frontiers in Network Physiology, 3, 1079070.
https://www.frontiersin.org/articles/10.3389/fnetp.2023.1079070/full
VCell 7.5 Release
2023-05-01. VCell 7.5 has been released. It features parameter estimation from the latest version of COPASI; model exchange via multiple standards, including, SBML, SEDML and OMEX; parameter scans in local client simulations; and hardening of unit handling including automatic unit transformations. Full specs available https://vcell.org/run-vcell-software.
VCell model of wild-type Bacillus subtilis spores
2023-03-01.VCell was used to demonstrate that expression of the 2Duf protein in wild-type Bacillus subtilis spores stabilizes inner membrane proteins and increases spore resistance to wet heat and hydrogen peroxide. Find links to the paper on our published models page.
VCell poster at BPS 2023 in San Diego
2023-02-20. The VCell team presented a poster at the Biophysical Society Annual Meeting in San Diego (https://www.biophysics.org/2023meeting#/).
A model of relationships between Ca2+ and YAP/TAZ signalling
2023-02-16. VCell collaborator Padmini Rangamani from University of California San Diego published a manuscript where a computational model of a mechanisms underlying divergent relationships between Ca2+ and YAP/TAZ signallingin was designed in VCell. The created system of ODEs was exported into MatLab for further simulations and processing.The model predicts context-dependent Ca2+ transient, CaMKII bistable response and frequency-dependent activation of LATS1/2 upstream regulators as mechanisms governing the Ca2+ -YAP/TAZ relationship.
Khalilimeybodi, A., Fraley, S. I., & Rangamani, P. (2023).
Mechanisms underlying divergent relationships between Ca2+ and YAP/TAZ signalling.
J Physiol. 601(3):483-515.
https://pubmed.ncbi.nlm.nih.gov/36463416/
One can check and download the VCell BioModel.
2023 Computational Cell Biology workshop
2023-02-13. The 24th Annual Computational Cell Biology workshop was sponsored by Cell Analysis and Modeling Center at the UConn Health on February 13-15th. It was an intense online event designed to enable cell biologists and biphysicists to develop mathematical models of their experimental systems.Keynote speakers included Jason Haugh, Pablo Iglesias and Ursula Kummer.
SpringSaLaD used for Coarse-Grained Molecular Modeling
SpringSaLaD software was used to model contractility kits, complexes in the cytoplasm of Dictyostelium cells consisting of myosin II and cortexillin I in the publication, Particle-based model of mechanosensory contractility kit assembly.
VCell and COPASI are used to study antibiotic biosynthesis
2022-09-28.VCell and COPASI were used together to study the kinetic behavior of antibiotic biosynthesis. The paper describes the same model designed in both COPASI (for parameter estimations) and VCell (for simulation reaction curves). Find links to the paper on our published models page.
VCell tutorial at ICSB 2022 in Berlin
2022-09-12. The VCell team will give a tutorial at the 21st International Conference on Systems Biology – the premier meeting on systems studies in biology, human evolution disease and planetary health. The meeting will take place in Berlin, Germany on October 8th-12th. The tutorial will be on October 9th at 12:30pm local time (https://www.icsb2022.berlin/satellite-events).
VCell is used for computational investigation of the dynamic control of cAMP signaling by PDE4 isoform types
2022-06-18. VCell collaborator Aurélie Carlier from Maastricht University published a manuscript on computational investigation of the dynamic control of cAMP signaling by PDE4 isoform types. The authors demonstrated that cAMP signaling is distinctly regulated by different PDE4 isoform types andthat this isoform-specificity should be considered in both computational and experimental follow-up studies to better define PDE4 enzymes as therapeutic targets in diseases in which cAMP signaling is aberrant.
Paes, D., Hermans, S., van den Hove, D., Vanmierlo, T., Prickaerts, J., Carlier, A. (2022). Biophys J., 121(14):2693-2711. doi: 10.1016/j.bpj.2022.06.019. Epub 2022 Jun 18.
https://pubmed.ncbi.nlm.nih.gov/35717559/
VCell biomodels are available at https://vcell.org/paes-2022
The 23rd Annual Workshop on Computational Cell Biology
2022-05-25. The 23rd Computational Cell Biology workshop took place at R. D. Berlin Center for Cell Analysis and Modeling (CCAM) on May 23-25th. For 3 days more than 40 online participants learnt how to use VCell, COPASI and SpringSalad software tools for modeling of cell biology systems. 16 participants participated in the “project track”, working individually with CCAM staff on developing their modeling projects.
New VCell Model of nutrients transporter network in plants
2022-04-14. A new compartmental VCell Model was used to explore nutrients transporter network in plants. Find links to the paper and BioModel on our published models page.
VCell is used in Nature biosynthesis paper
2022-02-10. VCell was used to analyze structure of a B12-dependent radical SAM enzyme in carbapenem biosynthesis in a new Nature paper (available on our published models page).
Teaching with VCell MathModels
2021-12-8. A discussion of how to use a VCell MathModel to teach mathematical modeling in cellular systems has been published by the Carlier lab with contributions from Cowan and Loew. Find links to the paper and public VCell models on our published models page.
New VCell Model of Nucleocytoplasmic Actin Dynamics
2021-11-24. A new VCell Model of actin dynamics was used to explore how phsophorylation of cofilin is involved in transmission of signals to the transcriptional machinery by depleting of nuclear G-actin. Find links to the paper on our published models page.
VCell Model of LINE-1 RNA lifecycle
2021-10-05. A new publication by Martin et al. describes a VCell model of the lifecyle of LINE-1 RNA elements in order to understand their activation and propagation. Find links to the paper and public VCell models on our published models page.
VCell Used to Validate new cAMP Model
2021-10-04. VCell was used to validate a new model for cAMP metabolism used to explore cAMP signaling in pulmonary microvascular endothelial cells. Find links to the paper and on our published models page.
VCell Model of ion transport in Kidney
2021-09-22. A new VCell model from the Carlier lab is combined with experiments to explore regulation of ion transport in the kidney proximal tubule. Find links to the paper and public VCell models on our published models page.
New model of FAK influence on YAP-TAZ
2021-09-10. VCell MathModels were used to explore how FAK activation influences YAP/TAZ translocation in a new publication by Eroume et al. in Biophysical Journal. Find links to the paper and public VCell models on our published models page.
New model of actin filament nucleation
2021-09-9. VCell models and COPASI parameter estimation tools were used to test molecular mechanisms for actin filament nucleation in a paper from the Loew and Pollard labs. Find links to the paper and public VCell models on our published models page.
New model of Sperm Calcium
2021-7-27. A new VCell model by Korobkin et al, explores calcium oscillations in mammalian sperm. COPASI software was also utilized for parameter estimation to fit the models. Find links to the paper and public VCell models on our published models page.
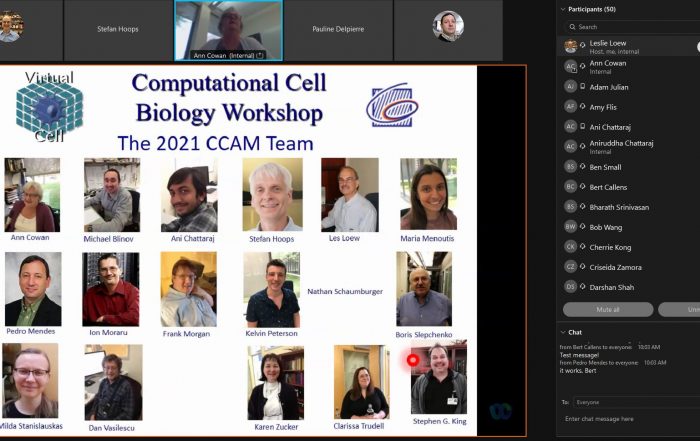
2021 Computational Cell Biology Workshop held June 21-23.
2021-6-24 CCAM’s annual Computational Cell Biology Workshop was again held online June 21-23, 2021. The workshop provided instruction to over 50 individuals from around the world on mathematical modeling techniques using VCell, COPASI and SpringSaLaD software. Fifteen of the participants worked closely with CCAM researchers to develop their own models related to their research projects. Another successful year for the modeling workshop! a combined workshop for VCell, COPASI, and SpringSaLaD modeling software. Thanks to all for a very successful course! Videos of the classes and presentations can be found on the CompCellBiol YouTube channel.
VCell model of YAP/TAZ
2021-5-08. A new VCell model from the Rangamani lab explores how YAP/TAZ signaling integrates inputs from different biophysical properties of the cell. Find links to the paper and public VCell models on our published models page.
VCell model of Axon Initial Segment
2021-4-05. A VCell model was used to assess the role of casein kinase 2 and ankyrin-G on sodium channel recruitment to the axon initial segment. Find links the citation on our published models page.
VCell model of Mitochondrial ATP production
2021-4-1. A new VCell model by Afzal et al, from the Lederer and Jafri labs explores how cristae geometry affects energy production in mitochondria Find links to the paper and public VCell models on our published models page.
VCell model of Mad signaling
2021-3-30. A new paper from the Inaba lab uses a VCell model to explore how pMad is asymmetrically partitioned to forming daughter nuclei during division of stem cells in the Drosophila ovary. Find links to the model and citation on our published models page.
VCell 7.4 released
2021-03-23.VCell 7.4 has been released. New in VCell 7.4 is the ability to preview geometries from the VCell database, vastly accelerated multiple trajectory stochastic simulations using the Gibson-Bruck solver, extension of unit display and automatic conversion to rate and assignment rules, support for external access to containerized versions of some VCell solvers with command line execution, as well as numerous additional improvements and bug fixes.
New VCell model of Ras signaling
2021-3-18. A new paper by Kerbai Eroume in the Carlier lab published in PLOS One explores how cell shape affects the signaling mechanisms that determine cell polarization. Find links to the model and citation on our published models page.
VCell 7.3 released
2020-11-30. Announcing the release of VCell 7.3! VCell 7.3 adds capabilities for multiple trajectories for non-spatial stochastic simulationss, interchange with the Open Modeling EXchange format (.omex), and automatic assignment of catalysts in the reaction diagram as well as numerous additional improvements and bug fixes.
VCell model of Ca-cAMP-PKA
2020-11-17. A VCell model was used to explore Ca2+-cAMP-PKA signalling circuits in a new publication from the Rangamani and Zhang laboratories. Find links to the model and citation on our published models page.
VCell model of agonist-induced signaling
2020-11-16. A new publication from the Lambert lab investigates the mechanisms that set the dynamic range of agonist-induced signaling using fluorescence imaging coupled with VCell modeling. Visit our published models page for links to the paper and to access the VCell model.
New VCell model of Ras signaling copy
2020-9-3. A new VCell model created incollaboration between the McCulloch and Zhang labs helped to demonstrate that a second phase of RhoA activation is required to maintain RhoA-mediated transcription after receptor signalling ends. Find links to the model and paper by Zhang et al. on our published models page.
VCell model of Ca2+ signaling in vascular smooth muscle
2020-10-20. A new publication from the Rangamani and Iyengar labs using VCell modeling to explore how cell shape affects calcium dynamics in vascular smooth muscle cells. Visit our published models page for links to the paper.
2020 CompCellBio Workshop
2020-08-05. The 2020 Computational Cell Biology Workshop was held as an online event Aug 3-5. Find videos of the key presentations here. Over 70 individuals participated overall; 16 individuals created models with VCell, COPASI and SpringSaLaD with help from CCAM team members in breakout rooms. A screen shot of one the opening day presentations serves in place of our traditional group photo this year.
VCell model of diffusion in axon initial segment
2020-07-10. A new publication analyzes fluorescence photobleaching experiments using VCell numerical simulations to explore mechanisms of for differentiating somatodendritic and axonal compartments. Visit our published models page for links to the paper.
VCell model of PLC/PKC signaling
2020-04-07. A new spatial model of the PLC/PKC pahtway created in VCell was used to explore chemotactic sensing in fibroblasts in a new publication by Nosbisch et al from the Haugh lab. Visit our published models page for links to the paper and the VCell model.
New VCell model of pattern formation system
2020-03-09. A new VCell model is used to explore how MYB proteins form an activator-inhibitor system that defines the spot pattern in mokeyflower petals. Visit our published models page for links to the paper and the VCell model.
VCell model of transport in plants
2020-01-30. A VCell model is used in a new paper by Dreyer and Michard to re-examine the concept of high- and low-affinity nutrient uptade systems in plants. Visit our published models page for links to the paper.
VCell 7.2 Released
2019-12-10. VCell 7.2 has been released. See the Release Notes for a full list of the many new features in VCell 7.2 including new functionality to store identifiers and text annotations for model components, a service linking VCell simulation with ImageJ image analysis functions, rate rules and assignment rules in ODE applications, improved SBML import along with several other improvements.
VCell model of cAMP signaling
2019-11-28. VCell was used to create a 2D spatial model of cAMP intracellular signaling in a new publication by Stone and colleagues. Visit our published models page for links to the paper.
20th Annual VCell Short Course
2019-6-18 CCAM hosted a Computational Cell Biology Workshop on June 24-26, 2019, a combined workshop for both VCell and COPASI modeling software. Ten scientists from the US, Netherlands and Germany traveled to work with the VCell and COPASI teams to construct models based on their own research interests. It was exciting to see VCell and COPASI models applied to such interesting cell biology projects. Thanks to all for a very successful course!
New model of GIV/girdin modulation of cyclic AMP signals
2019-06-13. A new Molecular Biology of the Cell publication from the Rangamani lab uses VCell and COPASI to develop a model to examine cross-talk between receptor tyrosine kinases and G proteins that regulate cAMP levels. Visit our published models page for links to the paper and to the VCell Biomodel.
VCell models aid in design of SH2 domain biosensors
2019-06-04. A new publication in Science Signaling from the Haugh and Rao labs describes the use of VCell modeling to assist in designing improved SH2 domain biosensors of EGFR phosphorylation. Visit our published models page for links to the paper and to the VCell Biomodel.
VCell model of RAF1 membrane dynamics published
2019-02-14. A VCell spatial model created to identify mechanisms regulating membrane abundance of the small Gprotein RAF1 at the plasma membrane has been published in Molecular Biology of the Cell. Visit our published models page for links to the paper and to the VCell Biomodel.
VCell model of voltage-sensing phosphatase specificity
2019-02-04. A new publication from the Hille lab uses a VCell model to reveal emergent properties of the behavior of voltage-sensitive phosphatases. Visit our published models page for links to the paper and to the VCell Biomodel.
New VCell analysis of FRAP experiments
2019-01-25. A new publication from Karvinen et al uses VCell models to analyze photobleaching experiments in their characterization of hydrogel properties. Visit our published models page for links to the paper.
VCell 7.1 released
17-11-2018. Announcing the release of VCell 7.1. VCell 7.1 adds the ability to explore existing models in the database and VCell functionality without registration. It has an improved model database info panel with model provenance, annotations, direct links to Pubmed and journal websites for models described in publications. These add to the new features for 2D kinematics to solve simulations with moving boundaries and the ability to use the COMSOL Multiphysics solver already in VCell 7.0.
New publication of VCell model of calcium dynamics in mossy fiber boutons
2018-07-10. A new Vcell spatial model of calcium influx, buffering and diffusion in mossy fiber boutons was published by Chamberland and co-authors in PNAS. Visit our published models page for links to the paper.
19th Annual VCell Short Course
2018-6-18 VCell hosted its 19th annual VCell Short Course on June 12-14, 2018. Ten scientists traveled to work with the VCell team to construct Virtual Cell models based on their own research interests. It was exciting to see the breadth of cell biological problems to which VCell models were applied. Thanks to all for a successful time!
VCell 7.0 with kinematics released
2018-3-15 Announcing the release of VCell 7.0. VCell 7 includes new 2D kinematics functionality for solving reaction diffusion equations within moving boundaries. To support models of cell motility and morphogenesis, this allows users to specify velocities of points, surfaces and volumes within the geometry. Other features in VCell 7.0 are methods for creating simulated fluorescence for direct comparison to microscopy data, the ability to use COMSOL Multiphysics solver (requires a local COMSOL license) and adaptive meshes for simulating with different spatial scales using EBChombo. VCell 7.0 has also been redesigned with new reusable modules and a cloud-hosted software development processes supporting external collaboration and extensibility (GitHub, Travis-Cl, DockerHub). VCell 7.0 also includes reproducible and portable server and solver deployments using container technologies.
The movie shows cell rotations in a model of actomyosin motility using the kinematics algorithm deployed in VCell 7.0; pseudocolors are myosin concentration. This model also includes cellular mechanics using an algorithm under development for future deployment in VCell. This work, and details of the model, are described in:
M. Nickaeen et al. (2017) A free-boundary model of a motile cell explains turning behavior. PLOS Computational Biology 13(11): e1005862. https://doi.org/10.1371/journal.pcbi.1005862
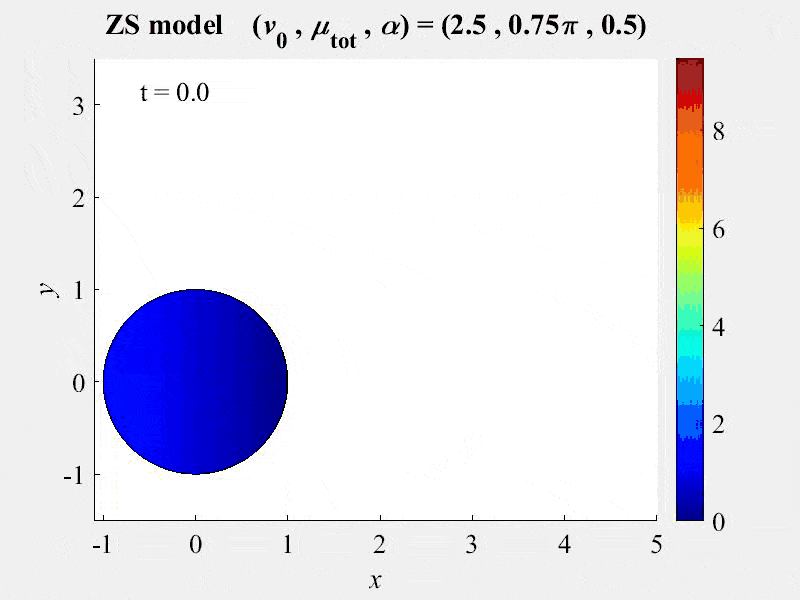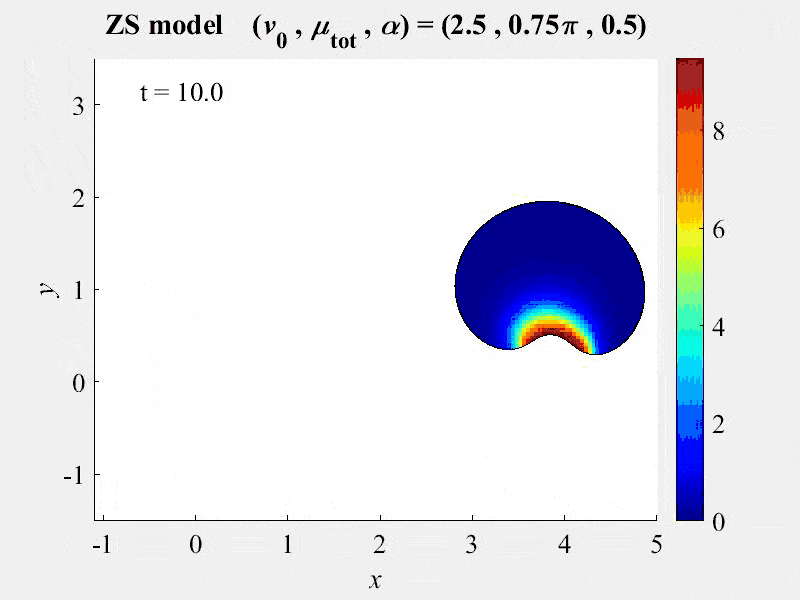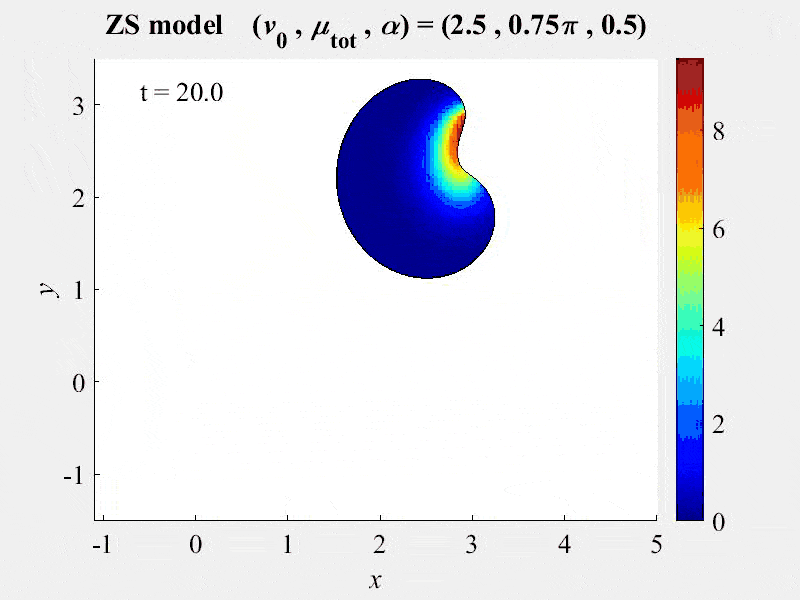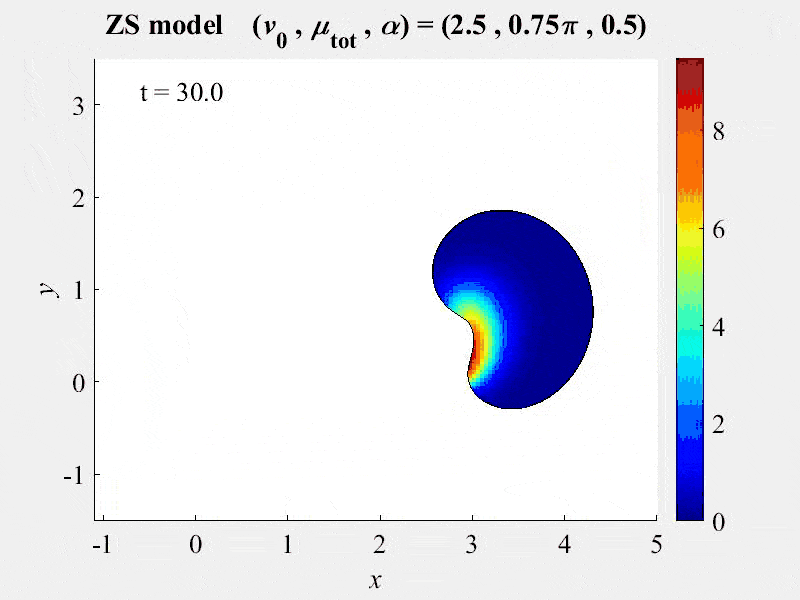
New publication assesses FRAP analysis methods using VCell VirtualFrap tool.
2018-03-13. A paper by Kinglsey et al assesses the effects of complex geometries on FRAP recovery curves. They develop a computational model of the FRAP processes to assess methods of analyzing FRAP experiments. They determined that the Vcell VirtualFRAP tool provides the best accuracy to measure diffusion coefficients. A link to the paper can be found on our published models page.
Leslie Loew awarded 2018 Biophysical Society Distinguished Service Award
2018-02-21. Leslie Loew received the Distinguished Service Award of the Biophysical Society at its 2017 annual meeting February 17-21. The award acknowledged his ongoing commitment to the Society and his dedicated service as Editor-in-Chief of Biophysical Journal. Congratulations to Les for this well-deserved award.

New publication on how cell shape information alters phenotype features VCell model
2017-12-15. A new publication from the Iyngar, Hone and He laboratories using Vcell among a number of other modeling strategies to explore how cell shape information can alter cellular phenotype via tension-independent mechansims. A link to the publication is found in our published models list.
SpringSaLaD version 2 released
11-30-2017. Version 2 of the SpringSaLaD software was released today. The primary new feature is the ability to directly build models from atomic coordinates in pdb files, using an interactive 3D viewer to compare the course-grained linked spheres representation in SpringSaLaD with the PDB structure.
New publication of novel free-boundary model of a moving cell
11-14-2017. A new publication, Nickaeen et al. 2017, from the VCell team describes a novel free-boundary model of actin -myosin contractility that couples force-balance and myosin transport equations. A previously developed mass-conservation algorithm originally developed for VCell to solve parabolic equations in moving domains with know kinematics was augmented by coupling with the FronTier front-tracking software and a segregated solver.
VCell model used to estimate diffusion coefficient in FRAP experiments.
2017-12-06. Simulations from a VCell model were used to estimate the diffusion coefficient of myosin II in fission yeast in a new publication from the Pollard laboratory. Visit our published models page for links to the paper and the public model.
New model of SH2 binding to EGFR published
2017-11-25. A new VCell model explores the effect of SH2 domain overexpression on the EGFR signaling pathway in a publication by Jadwin et al. The results suggest that signaling via SH2 domain binding is buffered over a wide range of concentrations. Explore the model from the listing on our Published Models page
Binding kinetics between VASP and Zyxin explored in VCell model
2017-8-27. In a new publication from Acevedo et al (2017) binding interactions between VASP and the cytoskeletal adapter protein Zyxin were explored using a VCell model to predict the population distribution of different molecular complexes based on different kinetic parameters. Find a link to the publication in our published models list.
New publication from Hille lab uses VCell model of GPCR signaling in neurons
2017-07-30. A new PNAS publication by Jung et al. is the latest in a series of VCell models published by the Hille lab. This new model explores the role of arrestin in regulating ERK activity during GPCR signaling. View the model structure through our list of published models.
New published model of the PLC/PKC pathway
2017-07-10. A new publication in Biophysical Journal by Mohan et al. uses a VCell model to define mechanisms for signal amplification in the PLC/PKC pathway during chemotaxis. Link to the publication and view model details from our Published Models listing.
Summer Research Projects
7-1-2017. CCAM welcomes several undergraduates and a graduate rotation student who are working on projects related to VCell this summer . Undergraduate students include Keeyan Ghoreshi, Anvin Thomas, Natalie de la Garrique and Shahan Kamal from UConn Storrs and Kevin Gaffney from the University of Oklahoma. Joe Masison is a new MD/PhD student from University of Maryland. Keeyan is working on the infrastructure for Sloppy Modeling projects, Shahan is modeling pathways using high-throuput data, Natalie is building Model Bricks, and Anvin is building VCell models for analyzing optogenetic experiments and developing general tools for assessing parameter identifiability in VCell. Kevin’s project involves coupling ImageJ technology with VCell to improve comparison of image data to simulation outputs, and Joe is working on an enhancement to SpringSaLaD, to allow coarse grained molecular models to be derived directly from atomic coordinates. In addition, a high school intern Nathan Schaumburger is helping to update VCell tutorials. We are excited by their excellent progress so far.
New VCell model of blood coagulation pathway
2017-6-30. A VCell model has been used to investigate activation of the contact pathway for blood coagulation, describing membrane-dependent reactions for activation of Factor XII and Factor XI in the presence of inhibitors. A link to the paper from Chelushkin et al., 2017 is found in our list of Published Models .
18th Annual VCell Short Course
VCell was pleased and honored to host its 18th annual VCell Short Course on June 12-14, 2017. Twelve national and international scientists traveled to work with VCell developers and administrators to construct Virtual Cell models based on their own, personal research interests. Thanks to all for a successful time!
VCell model of Plant and fungal transporters
2017-05-10. A VCell model has recently been published by Wittek et al. that describes the battle between fungal and plant sugar transporters. View model details from our Published Models listing.
New VCell model of kidney podocyte cytoskeleton
2017-03-10. A new model from the Iyengar and Loew laboratories examines how cytoskeletal dynamics effect local changes in the complex morphology of kidney podocytes. Link to the publication and view model details from our Published Models listing.
SpringSaLaD Update
Update released August 4, 2016. It fixes a minor bug in cluster size statistics. Go to CCAM Software to replace your current version.
New publication describes hybrid deterministic – stochastic spatial solver
A new publication in PLoS Computational Biology from Boris Slepchenko and coworkers describes the new hybrid deterministic stochastic spatial solver used in VCell. Schaff, J.C., F. Gao, Y. Li, I.L. Novak, and B.M. Slepchenko. 2016. Numerical Approach to Spatial Deterministic-Stochastic Models Arising in Cell Biology. PLoS Comput Biol. 12:e1005236. PMID 27959915
VCell 6.1 released to beta site
2016-10-14. A new version of VCell (6.1) was released to beta site. The new version replaces VCell 6.0 in beta, and enhances the new Rule-Based Modeling capabilities available in VCell. Advantages of Rule-Based Modeling in VCell are
- Specify rule-based models in a GUI, no scripting language required
- Rule-based models can span multiple compartments
- Reactions and rules can be mixed in one model
- Full support for rules in all VCell Application types (spatial, nonspatial, deterministic, stochastic).
- A set of rules can be simulated with Network-Free Simulator NFSim
- “Molecular Anchors” in rule-based models that keeps membrane-bound receptors attached to the membranes.
Visit the Download page to try the new VCell beta.
2016-09-15 A new publication in Bioinformatics by Jim Schaff and Michael Blinov describes the new Rule-based modeling features in VCell 6.0. See Schaff et al., 2016. Rule-based modeling with Virtual Cell. BioInformatrics 23:2880-2882, PMID 27497444
New VCell model of PDE role in AMPAR trafficking
2016-09-10. A new publication in JBC describes a new VCell model used by Song et al. to study the role of PDE1 and PDE2 in AMPA receptor trafficking in medium spiny neurons. Link to the publication and view model details from our Published Models listing.
New VCell model of dendritic spines
2016-08-30 A recent publication in PNAS describes a VCell model developed as collaboration between the Rangamini group at UC Davis and the Oster lab at UC Berkeley. The model couples biochemical signaling machinery with actin remodeling events in the dendritic spine; view the details on our Published Models list.
New VCell models of chemotactic networks
2016-07-27 A new publication from Sayak Bhattacharya and Pablo Iglesias at Johns Hopkins University describes the step-by-step construction of dynamical models of chemotactic networks using VCell (Bhattacharya and Iglesias 2016. Methods Mol. Biol. 1407:397, PMID 27271916). Publically available VCell models associated with the paper can be found on our list of published models here.
New VCell models of cAMP domains
2016-07-15 VCell models were used to test possible mechanisms for maintenance of subcellular microdomains of cAMP in cardiomyocytes in a recent publication in PLoS Computational Biology by Yang et al. Find the publication and details of the models on our Published Models listing.
VCell 3-day course June 20-22, 2016
12 investigators traveled to CCAM to work with the VCell team, developing a model relevant to their specific research project.
New VCell model of blood coagulation factors
2016 -06-09 Hysteresis-like binding of coagulation factors x/xa to procoagulant activated platelets and phospholipids results from multistep association and membrane-dependent multimerization (Podoplelova et al. 2016. Biochim Biophys Acta. 1858:1216-1227. PMID 26874201). Find the public BioModel here.
New VCell Model from the Hille lab
VCell models were used to quantitatively analyze FRET measurements were used to demonstrate that voltage-sensing phosphatases (VSPs) have catalytic activity for PIP3 (Keum et al. 2016. Phosphoinositide 5- and 3-phosphatase activities of a voltage-sensing phosphatase in living cells show identical voltage dependence. Proc Natl Acad Sci U S A. 10.1073/pnas.1606472113, PMID 27222577). Find the public VCell model here.
New VCell model of G-protein coupled receptors
2016-05-03 A new model of pancreatic beta cell G-protein coupled receptors and second messenger interactions was recently published in PLoS One by Fridlyand and Philipson. Go to our Published Models listing to link to the publication and view details of the model.
New published VCell Model of protease-activated receptor 2 signaling.
2016-03-01. A new publication describes a VCell model that evaluates the contributions of protein kinases and beta-arrestin to termination of protease-activated receptor 2 signaling. Visit the Published Models page for details and links to the publication.
New method for particle-based simulations with excluded volume.
2016-02-02 SpringSaLad (Springs, Sites, and Langevin Dynamics) is a new software that uses a course-grained approach to model biomolecules as a group of linked spherical sites with excluded volumes. The software user material is available here. Read about the method and software in Springsalad: A spatial, particle-based biochemical simulation platform with excluded volume. Michalski, P.J., and L.M. Loew. 2016 Biophys J. 110:523-529. PMID 26840718.
New VCell Model of phosphoinositide signaling
2016-06-08 A new VCell Model from the Hille lab describes the dynamics of phosphoinositide signaling in sympathetic neurons. Details of the model and links to the publication can be found on our Published Models listing.
New VCell model of PAR2 desensitization
2016-03-15. A new VCell model of the mechanisms and kinetics of desensitization of the protease-activated receptor-2 (PAR2), a Gq-PCR, was published by Jung et al. in the Journal of General Physiology. Visit the Published Models listing to view the model details and link to the publication.
2016-02-02 Paul Michalski and Les Loew describe their new Spring SaLaD sofware for spatial, stochastic, particle-based modeling of biochemical systems at the mesoscale. See the publication Michalski and Loew, 2016. SpringSaLaD: A Spatial, Particle-Based Biochemical Simulation Platform with Excluded Volume. Biophysical Journal 110:523-529. PMID 26840718